All published articles of this journal are available on ScienceDirect.
Current Status of Computational Intelligence Applications in Dermatological Clinical Practice
Abstract
Background:
The yeast infections are increasingly frequent and the correct diagnosis consists of the identification of the yeast fungus, which in our case we are going to refer to the different species of Candida. The prescription of a broad-spectrum antifungal without taking into account the etiological agent, leads to an increase in the resistance to these treatments.
Objective:
The objective of this work is to differentiate Candida albicans from other Candida species (Candida spp.) By means of digital images obtained from the optical microscope.
Material and Methods:
It has reviewed about 100 photographs from patients in our consultations.
In this study we will use the microscopic images of the Candida variety to be processed later with the Octave programming language and its image processing package (image-2.8.0).
Results and Discussion:
This system is able to differentiate Candida albicans from the other varieties of Candida such as C. parapsilosis, C. krusei, and C. kefyr with accuracy.
The candida identifier application, which was designed and programmed in Octave, allows identification of candida species by locating certain geometric descriptors, such as the centroid and the surfaces of circular objects within the images. The program was highly effective for the diagnosis of Candida spp. So, we got a sensitivity and specificity above 90% with the images used.
Conclusion:
The results that we obtain from the Candida spp. identifier system that opens the way to be able to work with images obtained from the optical microscope.
1. INTRODUCTION
Intelligent computer systems already provide support to healthcare professionals.
Computational intelligence has been widely researched and applied owing to its ability to cope with large amounts of clinical data and uncertain information [1]. Computational intelligence uses algorithms based on biological data, primarily on neuronal functioning. The three pillars on which computational intelligence is based are neural networks, genetic algorithms, and fuzzy systems [2]. Neural networks are algorithms employed for function approximation or classification problems. They include supervised, unsupervised, and reinforcement learning [3].
Constant progress is being made on computational intelligence to improve the prevention, diagnosis, and treatment of diseases in general, and mycosis in particular [4].
The yeast Candida albicans is the most prevalent pathogenic Candida species (spp.), although the increase in immunocompromised patients has been accompanied by an increase in the diversity of pathogenic strains found as etiological agents of fungal infections [5, 6].
The most typically used method to identify Candida spp. is the chromogenic culture medium. An analysis of the growth facilitates the determination of the purity of the colonies, and the identification of Candida spp [7].
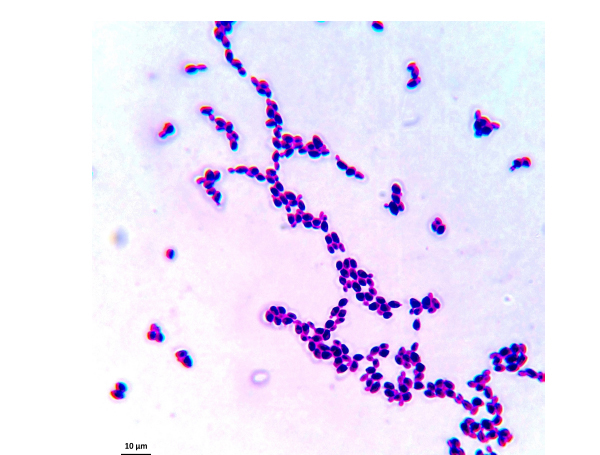
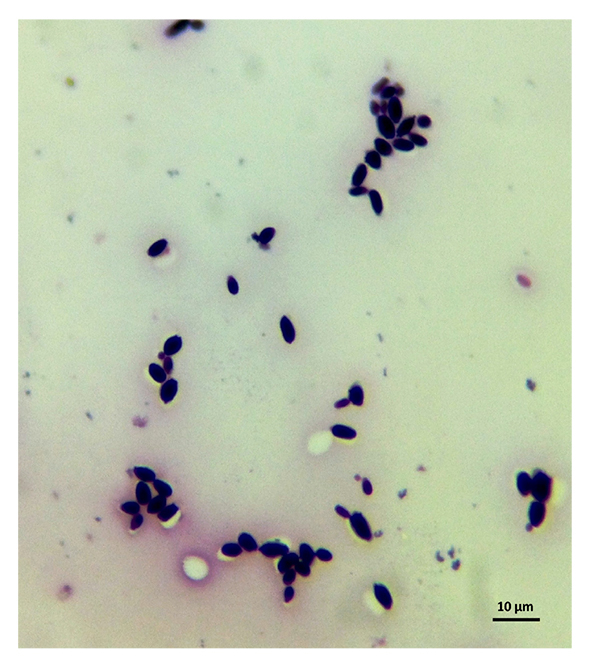
The samples used by us are from this culture medium and were later imaged under an optical microscope. After reviewing several images, we have selected C. albicans, C. parapsilosis, C. krusei, and C. kefyr for our project. The images used in the elaboration of the program correspond to Figs. (1-7).
2. MATERIALS AND METHODS
2.1. Material
Approximately 100 photographs primarily from the author’s patients were reviewed.
In this study, microscopic images of the Candida species were used and subsequently processed using the Octave programming language and its image processing package (image-2.8.0). The interface was obtained through Electron's libraries, based on JavaScript and HTML.
2.2. Description of the Method
Development of programs for the detection of Candida species
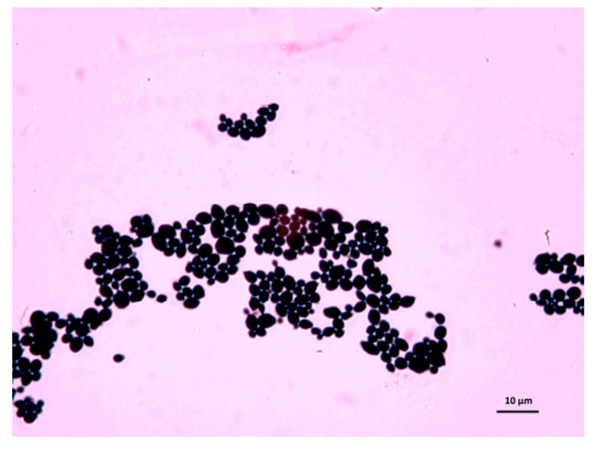
As the characteristics of the different species of Candida are known, we applied digital image processing with Octave. We performed an adaptation of the diagnostic methods of the Candida and their subsequent study under a microscope for the classification of the different species Most of these methods have typical concepts, such as the variety of colors, asymmetry in their internal forms, and abrupt and irregular edges, and are used to identify elements of the said fungi under the optical microscope. These are the primary concepts that are analyzed in the programs performed, and are referred to as the “Candida Identifier.”
3. OBJECTIVES
- The objective of this work is to differentiate Candida albicans from other Candida spp. with an accuracy of approximately 70%.
This general objective is divided into more specific objectives as follows:
- Establish the spaces of color and form that will be used in this work.
- Define the necessary programs, in Octave environment, such that they can recognize the different species of Candida
- Develop a rapid, easy, and economical technique for identifying Candida spp. that is easily implementable and manageable even by primary care physicians.
4. RESULTS AND DISCUSSION
(1) The identification algorithm was performed using the following steps:
- Load the images
- Go from RGB to grayscale
- Binarization of the image
- Tag pixel regions to establish neighborly relations
- Perform geometric measurements to select descriptors
- Pattern determination
- Choice of patterns to differentiate the species of Candida (Candida spp.)
(2) Later, we will perform the stages of the model comprising the following:
(A) Identification and training criteria
(B) Testing
(C) Validation
(D) Implementation of the algorithm through Octave and program development
(E) Interface design
4.1. Identification and Training Criteria
Subsequently, with part of the program code, we seek to identify the circularity of each element, which is stored in the circularities variable. The results are weighted with an arbitrary threshold to define the number of circular elements of the images, which is stored in the variable roundObjects. These characteristics allow us to differentiate the species studied. For example, C. albicans exhibits circular elements resembling C. kefyr. Meanwhile, C. krusei exhibits more linear elements.
To calculate of the circularity of rounded objects we use the follow program code in Table 1.
Table 1.
| circularities = allPerims.^ 2./ (4*pi*allAreas); Weighting to discriminate circular components: roundObjects = find(circularities<4); |
4.2. Test Three Times Using the Images
In the testing stage, it was possible to verify the application from the original image to grayscale and subsequently to its binary form to determine its geometric properties through the Octave “regionprops” function. This function returns the patterns that characterize each type of fungus, thus allowing for the effective identification of the images. The different steps to be taken are: First of all, we have the original image. The second step is to pass it to grayscale image and finally we get the binarized image.
4.3. To Determine Our Model Behavior When we Apply it to Candida sp., we Constructed a Confusion Matrix (Table 2 &3)
| Comparison | Identifier Value | ||||
|---|---|---|---|---|---|
| Real Value | x/x | C. albicans | C. krusei | C. kefyr | C. parapsilosis |
| C. albicans | 100 | 0 | 0 | 0 | |
| C. krusei | 0 | 90 | 10 | 0 | |
| C. kefyr | 0 | 10 | 90 | 0 | |
| C. parapsilosis | 0 | 0 | 0 | 100 | |
4.4. Implementation of the Algorithm Through Octave and Development of the “Candydos” Program
The program has implemented the image recognition algorithm, using the leeImage function. In its first line, the image library of Octave is loaded. This library contains the classes and functions that allow for the processing of mushroom images (Table 4)
4.5. Interface from Database of Java Script Libraries
Finally, as Octave does not have a method to generate the GUI, the interface has been implemented through the Electron framework, which is developed in JavaScript, and HTML (Table 5).
Table 4.
| function [identificador] = leeImagen(file) pkg image load; % This part reads the file of the image that will be classified by the% recognition program. RGB = imread (file); % The RGB image is passed in grayscale, that is, the original image with its component% Red, Green, and Blue I = rgb2gray (RGB); % In this stage, the image that is in ray scale is binarized. This operation passes% first from the format uint8 to double. Having this format, the scale of gray% is compared with the threshold 0.2. Values above 0.2 become! (white) and below% 0 (black). BW = im2bw(I,0.2); % The image is displayed % imshow (BW) % The bwlabel command labels the binary image, such that it establishes contiguous (neighboring) zones to determine the regions. abeledImage = bwlabel (BW); % The regionprops function allows for the determination of the geometric descriptors of the figures obtained, after binarizing the images. The areas are found, as well as the perimeters that are essential% to initially identify the circular regions. This descriptor allows the% fungi types to be differentiated. Thus, for example, the circular area of a Candida% albicans fungus differentiates it from a parapsilosis fungus%. measurements = regionprops (labeledImage, 'Area', 'Perimeter'); allAreas = [measurements.Area]; allPerims = [measurements.Perimeter]; circularities = allPerims. ^ 2./ (4 * pi * allAreas); blobMeasurements = regionprops (labeledImage, 'BoundingBox', 'Area'); allBlobAreas = [blobMeasurements.Area]; % Find objects that have “round” values of circularities. roundObjects = find (circularities <4); % Whatever value you want. % Compute new binary image with only the round objects in it. binaryImage = ismember (labeledImage, roundObjects)> 0; identificador = find(circularities < 4 & allBlobAreas > 10000); %figure(2) %imshow(binaryImage); Endfunction % In this stage, the images that are being loaded to the application% are simply classified according to the identifier that has been defined for the fungi in the function. % clear all; clc; # not a function file: pkg image load; file = 'ima.jpg'; [identifier] = readImage (file); if length (identifier) == 1 && (identifier <10) disp (“The fungus is Candida parapsilosis”) elseif length (identifier) == 1 && (identifier> 20) && (identifier <30) disp (“The fungus is Candida albicans”) elseif length (identifier> 100) disp (“The fungus is Candida kefyR”) else disp (“The fungus is Candida krusei”) endif fflush.stdout () |
Table 5.
| <! DOCTYPE html> <html> <head> <meta charset = “UTF-8”> <title> Mushroom Recognition </ title> </ head> <style> .body { background: # 000; color: #fff; overflow: hidden; } </style> <body class=”body”> <input type=”file” onchange=”onImageSelected(event)”/> <button type=”button” id=”htmlButtonPress” onclick=”launchPython()” >Clasifica</button> <img id=”myimage” width=600 height=400 > <canvas id=”myCanvas”/> <section> <script> var buffer = require('buffer'); var path = require('path'); var fs = require('fs'); // Here, the image that is identified in the Canvas is loaded as “myimage. function onImageSelected(event) { var selectedFile = event.target.files[0]; var reader = new FileReader(); var imgtag = document.getElementById(“myimage”); reader.onload = function(event) { imgtag.src = event.target.result; var imagen = imgtag.src; var data = imagen.replace(/^data:image\/\w+;base64,/, “"); // console.log(“La data es:”+data) var buf = new Buffer(data, 'base64'); fs.writeFile('ima.jpg', buf, (error) => {/* handle error*/}); }; reader.readAsDataURL(selectedFile); } const PowerShell = require(“powershell”); // Here, the communication with Octave is made so that it executes the programs. function launchPython() { let ps = new PowerShell('C:/Octave/octave-4.4.1/bin/octave-cli -qf demos.m'); ps.on(“error”, err => { console.error(err); }); // Stdout ps.on(“output”, data => { console.log(data); const Par = document.createElement('p') Par.textContent = data; document.querySelector('body').appendChild(Par); }); } </script> // Modules to control application life and create native browser window const {app, BrowserWindow} = require('electron') // Keep a global reference of the window object. If you do not, the window will // be closed automatically when the JavaScript object is garbage collected. let mainWindow function createWindow () { // Create the browser window. mainWindow = new BrowserWindow({width: 600, height: 700}) // and load the index.html of the app. mainWindow.loadFile('index.html') // Open the DevTools. // mainWindow.webContents.openDevTools() // Emitted when the window is closed. mainWindow.on('closed', function () { // Dereference the window object, typically you would store windows // in an array if your app supports multi windows, this is the time // when you should delete the corresponding element. mainWindow = null }) } // This method will be called when Electron has finished // initialization and is ready to create browser windows. // Some APIs can only be used after this event occurs. app.on('ready', createWindow) // Quit when all windows are closed. app.on('window-all-closed', function () { // On OS X, it is typical for applications and their menu bar // to stay active until the user quits explicitly with Cmd + Q if (process.platform!== 'darwin') { app.quit() } }) app.on('activate', function () { // On OS X, it is typical to re-create a window in the app when the // dock icon is clicked and there are no other windows open. if (mainWindow === null) { createWindow() } }) // In this file, you can include the remainder of your app's specific primary process // code. You can also place them in separate files and require them here. See translate video of program operation in Appendix 1. |

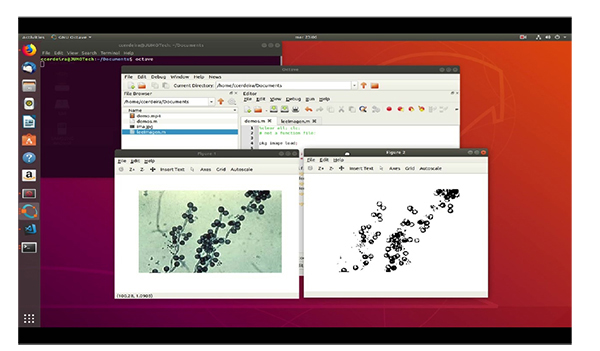
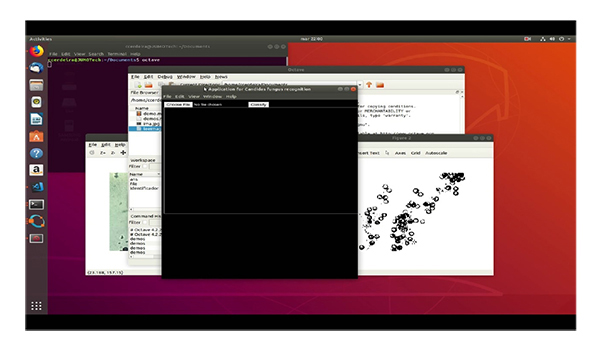
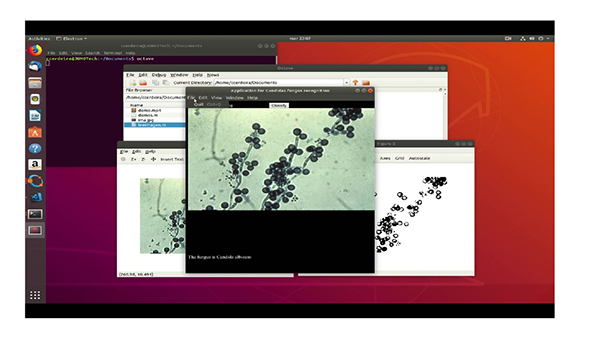
5. DISCUSSION
A major difficulty was in obtaining a large sample and images of the Candida spp. used with similar characteristics, because the time of planting and the technique vary according to its shape and size. The system cannot change the size of the images during the entire process [9].
Another difficulty that also influences the classification of the image is the image quality [10, 11].
The more the noise or the poorer the image quality, the less precision the edge mapping process will have [12].
Further, the less accuracy on the border mapping process, the worse is the performance on the classification process [13].
In the literature, we did not find models that were used to recognize Candida spp. However, we found references bout the artificial neural networks to calculate antifungal activity against C. albicans. Furthermore, there are references to other cutaneous diseases [14-16] such as melanoma [17].
Hence, we established a goal. The objective was to reach a diagnostic accuracy of at least 80%. The model proposed by Manousaki et al. [18] provided an accuracy of 89.4%. The model for pre-diagnostic digital imaging developed by Christensen et al. [19] presented an accuracy of 77%. Our model is superior in accuracy to those mentioned above.
The final prediction model developed exhibits an accuracy of almost 90% and will thereby represent an improvement in diagnostic accuracy. Despite the different appearances and qualities of the images in the image database, we could develop a prediction model with accuracy better than that expected from an experienced dermatologist [20].
The algorithms are thereby stable and not dependent on the quality of the image |under inspection. These new tools can also be utilized in telemedicine, where images can be uploaded to an automated web-based database and subsequently analyzed [21].
CONCLUSION
We designed and programmed the “Candida identifier application” in Octave. It allowed the identification of the Candida species by locating certain geometric descriptors, such as the centroid and the surfaces of the circular objects that comprise the images. This system could differentiate the C. albicans from other varieties of Candida such as C. Kefyr, C. parapsilosis, and C. krusei, with accuracy. The analysis by the system of these descriptors allowed us to identify the Candida species in more than 90% of the cases. The sensitivity and specificity that were above 90% indicated a high diagnostic efficacy that was rather large to be the first version of a diagnostic program.
The application was highly precise in the identification based on the forms; however, it did not exhibit the same features in color-based identification. This element typically presents variations in the data, thus rendering identification difficult. We should acquire more copious databases to implement more precise identification methods, such as deep learning algorithms for images; however, the recognition of information in three different areas was satisfactory, as it allowed us to determine and classify injuries within the medical margins.
Finally, we emphasize that the objectives set at the beginning of the project were achieved. The results that we obtained from the “Candida identifier system” paved the way for handling images obtained from an optical microscope. This implies a wider range of materials can be used and specialized personnel economy (mainly in developing countries), and better service from the patient's perspective.
AUTHORS' CONTRIBUTORS
“Candydos” program. CRC designed and implemented the algorithm for “candydos” through Octave, as well as developed its program.
JLGC has contributed to create the algorithms of the program
RA has provided and selected Candida spp.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study was approved by the Ethics and Research Committee of Hospital General Manuel Gea Gonzalez, Estado de México, Mexico. (Approval Number: OF /No 145001022 151/CCEEIS/016/2018).
HUMAN AND ANIMAL RIGHTS
No Animals were used in this research. All human research procedures followed were in accordance with the ethical standards of the committee responsible for human experimentation (institutional and national), and with the Helsinki Declaration of 1975, as revised in 2013.
CONSENT FOR PUBLICATION
Written informed consent was obtained from patients for the use of the fungal isolates and the publication of the cases details.
AVAILABILITY OF DATA AND MATERIAL
The data that support the findings of this study are available from the corresponding author [C.C] upon request.
FUNDING
The authors received no financial support for the research, authorship, and/or publication of this article.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
Declared none.
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publishers Website along with the published article.
APPENDIX 1
DESCRIPTION OF THE VIDEO CONTENT
The video includes the creation of the algorithm, and the diagnosis and operation of the program:
First, “%” passes the RGB image in grayscale, that is, the original image with its “%” red, green, and blue components (binarization of the image).
Subsequently, “%” classifies the images that are loaded to the application% according to the identifier that has been defined for the fungi in the “readImage” function
Finally, how the different species of Candida are presented and how the program identifies them with an accuracy of at least 90% are observed.
Steps of the video
1
00: 00: 02,193 -> 00: 00: 03,003
<i> Program code showing the results </ i>
2
00: 00: 04,193 -> 00: 00: 06,003
<i> Code of the program that analyzes the images </ i>
3
00: 00: 07,193 -> 00: 00: 11,003
<i> Save and play the program </ i>
4
00: 00: 20,193 -> 00: 00: 24,003
<i> Real and binarized image </ i>
5
00: 00: 25,193 -> 00: 00: 28,503
<i> Command window: The fungus is Candida kefyr </ i>
6
00: 00: 44,193 -> 00: 00: 46,003
<i> Graphic interface </ i>
7
00: 00: 46,993 -> 00: 00: 49,503
<i> Choose file </ i>
8
00: 00: 52,073 -> 00: 00: 56,003
<i> Classify: The fungus is Candida albicans </ i>
9
00: 00: 59,193 -> 00: 01: 02,503
<i> Choose file </ i>
10
00: 01: 04,033 -> 00: 01: 07,021
<i> Classify: The fungus is Candida kefyr </ i>
11
00: 01: 08,193 -> 00: 01: 09,803
<i> Choose file </ i>
12
00: 01: 14,173 -> 00: 01: 17,303
<i> Classify: The fungus is Candida parapsilosis </ i>


